Zusammenfassung: Letzte Sitzung
- Parameterschätzung: Wahrscheinlichkeitsparameter einer Binomialverteilung
- Beta Verteilungen
- Grid Approximation
Bayes’ Theorem
\[ P(\theta|Data) = \frac{ P(Data|\theta) * P(\theta) } {P(Data)} \] oder ohne Normalisierungskonstante:
\[ P(\theta|Data) \propto P(Data|\theta) * P(\theta) \]
Parameterschätzung: Parameter einer Binomialverteilung
Die Binomial Likelihood ist gegeben durch
\[ P(x = k) = {n \choose k} \theta^k (1-\theta)^{n-k} \]
wins <- 6
games <- 9
Maximum Likelihood Schätzung
theta <- wins/games
theta
[1] 0.6666667tibble(x = seq(from = 0, to = 1, by = .01)) %>%
mutate(density = dbinom(6, 9, x)) %>%
ggplot(aes(x = x, ymin = 0, ymax = density)) +
geom_ribbon(size = 0, alpha = 1/4, fill = "steelblue") +
geom_vline(xintercept = theta, linetype = 2, size = 1.2) +
scale_y_continuous(NULL, breaks = NULL) +
coord_cartesian(xlim = c(0, 1)) +
xlab("Wahrscheinlichkeit") +
theme(panel.grid = element_blank(),
legend.position = "none")

n_points <- 100
theta_grid <- seq(from = 0 , to = 1 , length.out = n_points)
likelihood <- dbinom(wins , size = games , prob = theta_grid)
prior <- dbeta(x = theta_grid, shape1 = 4, shape2 = 4)
unstandardized_posterior <- likelihood * prior
posterior <- unstandardized_posterior / sum(unstandardized_posterior)
d <- tibble(theta_grid, prior, likelihood, posterior)
d %>%
pivot_longer(-theta_grid, names_to = "distribution", values_to = "density") %>%
mutate(distribution = as_factor(distribution)) %>%
ggplot(aes(theta_grid, density, color = distribution)) +
geom_line(size = 1.5) +
geom_vline(xintercept = 9/10, linetype = "dashed") +
scale_color_viridis_d(end = 0.8) +
xlab("Theta Werte") +
ylab("") +
facet_wrap(~distribution, scales = "free_y") +
theme_bw() +
theme(legend.position = "none")
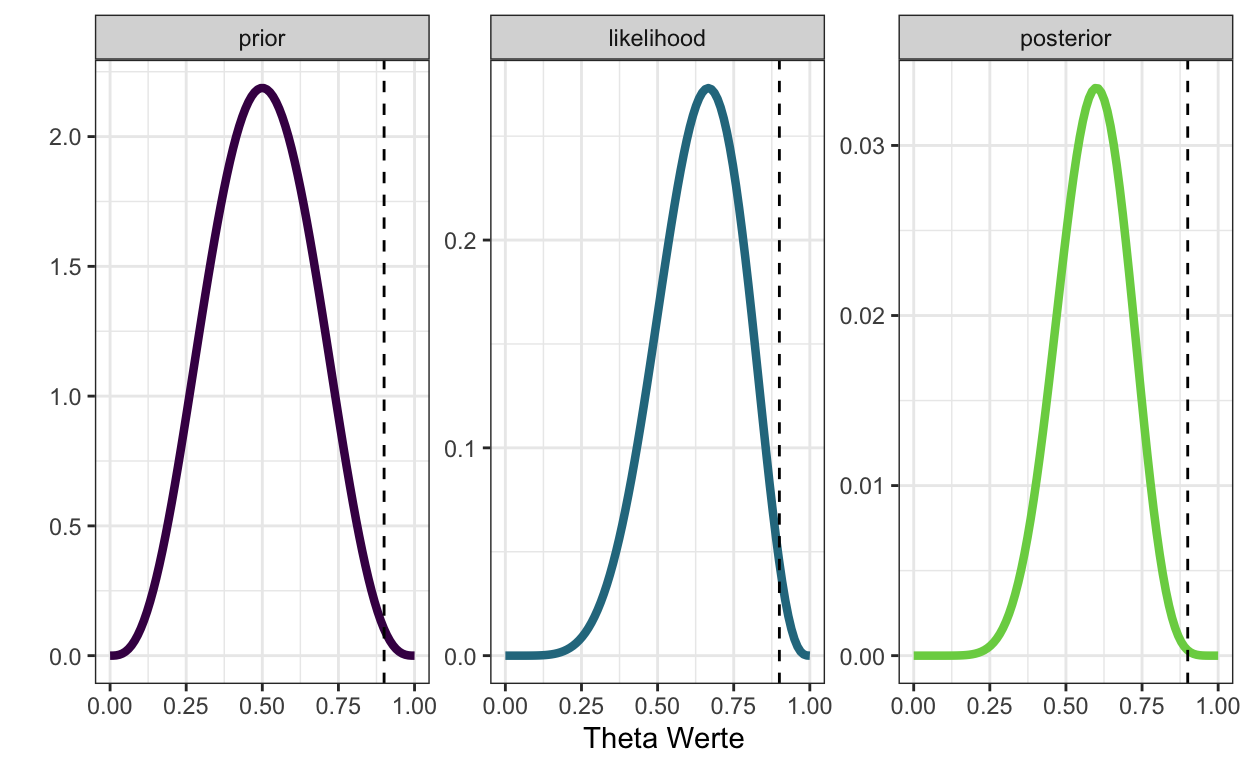
Posterior Verteilungen zusammenfassen
Samples aus dem Posterior ziehen:
n_samples <- 1e4
set.seed(3) # wegen Reproduzierbarkeit
d %>%
paged_table(options = list(rows.print = 6))
samples <-
d %>%
slice_sample(n = n_samples, weight_by = posterior, replace = TRUE) %>%
mutate(sample_number = 1:n())
samples %>%
paged_table(options = list(rows.print = 6))
samples %>%
ggplot(aes(x = sample_number, y = theta_grid)) +
geom_point(alpha = 1/10) +
scale_y_continuous("Erfolgswahrscheinlichkeit", limits = c(0, 1)) +
xlab("sample number")

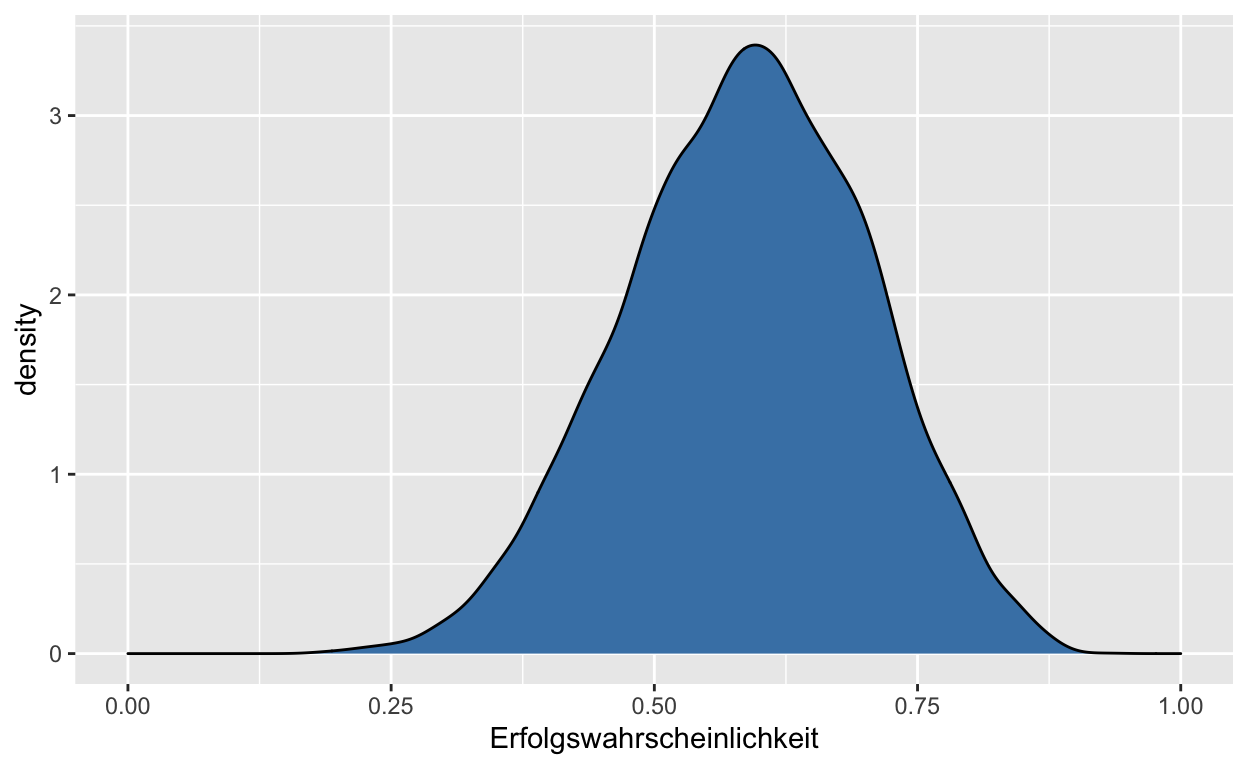
Intervalle mit definierten Endpunkten
Posterior Approximation
sum(posterior[theta_grid > 0.5])
[1] 0.7728132# A tibble: 1 x 1
sum
<dbl>
1 0.773Intervalle mit definierten Endpunkten
Posterior Samples
samples %>% #<<
filter(theta_grid > .5) %>% #<<
summarise(sum = n() / n_samples) #<<
# A tibble: 1 x 1
sum
<dbl>
1 0.773samples %>%
count(theta_grid > .5) %>%
mutate(probability = n / sum(n))
# A tibble: 2 x 3
`theta_grid > 0.5` n probability
<lgl> <int> <dbl>
1 FALSE 2273 0.227
2 TRUE 7727 0.773Intervalle mit definierter Masse
Quantile
y ymin ymax .width .point .interval
1 0.5959596 0.5050505 0.6666667 0.5 median qi y ymin ymax .width .point .interval
1 0.5959596 0.5050505 0.6666667 0.50 median qi
2 0.5959596 0.4343434 0.7373737 0.80 median qi
3 0.5959596 0.2929293 0.8484848 0.99 median qiHighest Posterior Density (HPDI)
mode_hdi(samples$theta_grid, .width = .5)
y ymin ymax .width .point .interval
1 0.5961325 0.5151515 0.6666667 0.5 mode hdihdi(samples$theta_grid, .width = .5)
[,1] [,2]
[1,] 0.5151515 0.6666667library(patchwork)
p1 <-
d %>%
ggplot(aes(x = theta_grid)) +
# check out our sweet `qi()` indexing
geom_ribbon(data = d %>% filter(theta_grid > qi(samples$theta_grid, .width = .5)[1] &
theta_grid < qi(samples$theta_grid, .width = .5)[2]),
aes(ymin = 0, ymax = posterior),
fill = "grey75") +
geom_line(aes(y = posterior)) +
labs(subtitle = "50% Percentile Interval",
x = "Erfolgswahrscheinlichkeit",
y = "density")
p2 <-
d %>%
ggplot(aes(x = theta_grid)) +
geom_ribbon(data = . %>% filter(theta_grid > hdi(samples$theta_grid, .width = .5)[1] &
theta_grid < hdi(samples$theta_grid, .width = .5)[2]),
aes(ymin = 0, ymax = posterior),
fill = "grey75") +
geom_line(aes(y = posterior)) +
labs(subtitle = "50% HPDI",
x = "Erfolgswahrscheinlichkeit",
y = "density")
p1 | p2

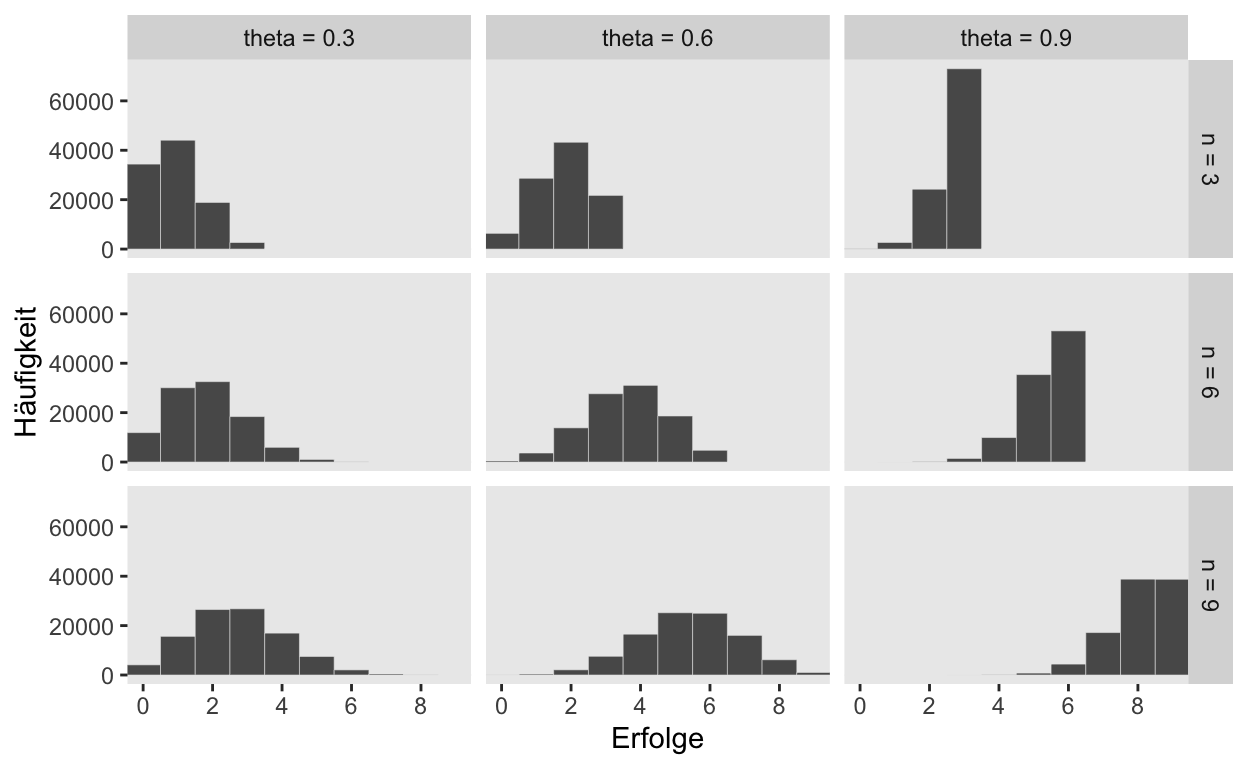
Sampling: Simulation

n_draws <- 1e5
simulate_binom <- function(n, probability) {
set.seed(3)
rbinom(n_draws, size = n, prob = probability)
}
d <-
crossing(n = c(3, 6, 9),
probability = c(.3, .6, .9)) %>%
mutate(draws = map2(n, probability, simulate_binom)) %>%
ungroup() %>%
mutate(n = str_c("n = ", n),
probability = str_c("theta = ", probability)) %>%
unnest(draws)
d %>%
ggplot(aes(x = draws)) +
geom_histogram(binwidth = 1, center = 0,
color = "grey92", size = 1/10) +
scale_x_continuous("Erfolge",
breaks = seq(from = 0, to = 9, by = 2)) +
ylab("Häufigkeit") +
coord_cartesian(xlim = c(0, 9)) +
theme(panel.grid = element_blank()) +
facet_grid(n ~ probability)
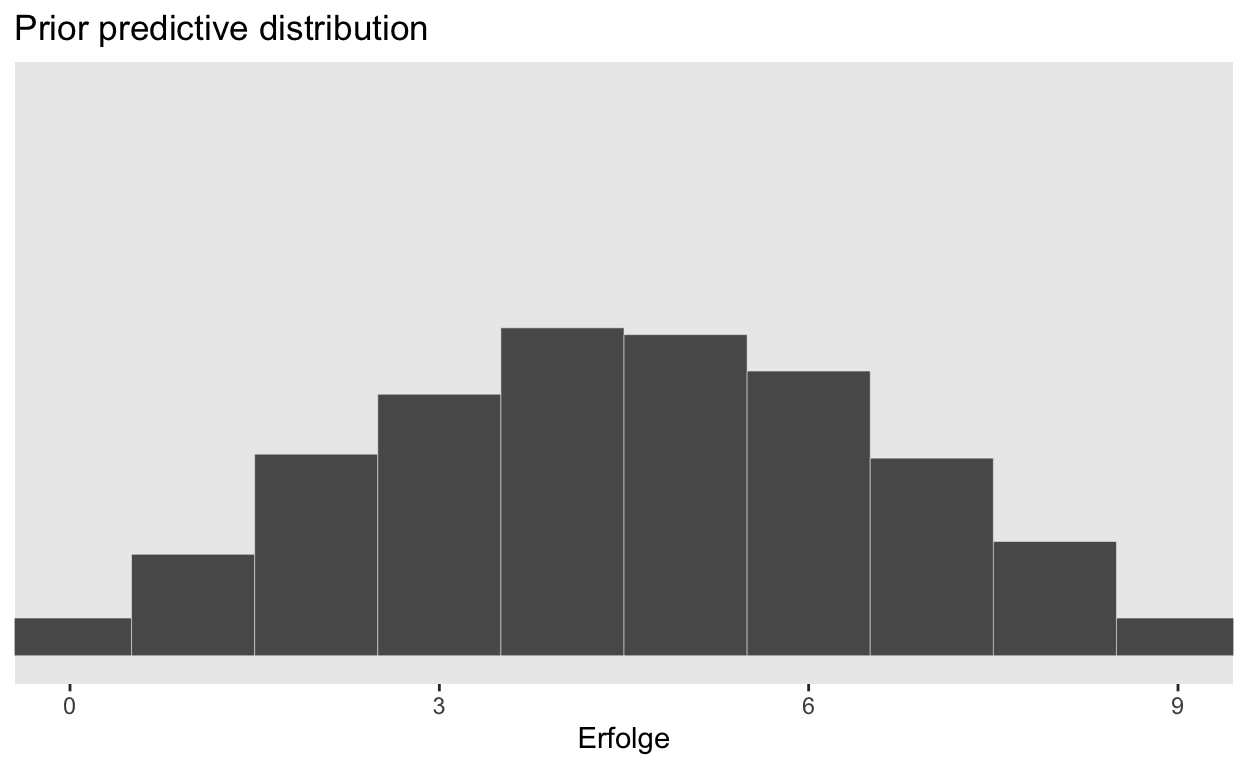
Prior Predictive Distribution
Posterior Predictive Distribution

Graphical model
Binomial Model
Grafische Darstellung des Generativen Modells:
Gaussian model
Grafische Darstellung eines Generativen Modells mit normalverteilten Daten:
hist(y, col = 'skyblue3', breaks = 10)
Posterior Inference
// The input data is a vector 'y' of length 'N'.
data {
int<lower=0> N;
vector[N] y;
}
// The parameters accepted by the model. Our model
// accepts two parameters 'mu' and 'sigma'.
parameters {
real mu;
real<lower=0> sigma;
}
// The model to be estimated. We model the output
// 'y' to be normally distributed with mean 'mu'
// and standard deviation 'sigma'.
model {
mu ~ normal(120, 5);
sigma ~ uniform(1, 10);
y ~ normal(mu, sigma);
}priors <- set_prior("normal(120, 5)", class = "Intercept") +
set_prior("uniform(1, 10)", class = "sigma")
fit <- brm(y ~ 1,
family = gaussian,
prior = priors,
data = d,
cores = parallel::detectCores())
brms Model
get_prior(y ~ 1,
family = gaussian,
data = d)
prior class coef group resp dpar nlpar bound
student_t(3, 123, 4.4) Intercept
student_t(3, 0, 4.4) sigma
source
default
defaultm <- brm(y ~ 1,
family = gaussian,
prior = priors,
data = d,
cores = parallel::detectCores(),
file = "models/model_1")
summary(m)
plot(m)

m %>%
spread_draws(b_Intercept) %>%
median_qi(.width = c(.50, .80, .95)) %>%
kableExtra::kbl()
| b_Intercept | .lower | .upper | .width | .point | .interval |
|---|---|---|---|---|---|
| 121.9501 | 121.4131 | 122.4658 | 0.50 | median | qi |
| 121.9501 | 120.9458 | 122.9398 | 0.80 | median | qi |
| 121.9501 | 120.4171 | 123.4596 | 0.95 | median | qi |
m %>%
spread_draws(b_Intercept) %>%
ggplot(aes(x = b_Intercept)) +
stat_halfeye(.width = c(.50, .80, .95))
