Daten bearbeiten und zusammenfassen
Daten aus Verhaltensexperiments bearbeiten und zusammenfassen, Datenpunkte identifizieren.
Ob eine Variable als factor definiert ist, wird als Attribut gespeichert. Attribute werden aber in einem .csv. File nicht mitgespeichert; deshalb müssen wir die Gruppierungsvariablen wieder als factor definieren.
data <- data |>
mutate_if(is.character, as.factor)glimpse(data)Rows: 1,440
Columns: 9
$ trial <dbl> 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17…
$ ID <fct> JH, JH, JH, JH, JH, JH, JH, JH, JH, JH, JH, JH, JH, JH, JH, …
$ cue <fct> right, right, none, none, left, none, none, left, left, none…
$ direction <fct> right, right, right, right, left, right, left, left, right, …
$ response <dbl> 1, 1, 0, 1, 1, 1, 1, 0, 0, 1, 0, 0, 0, 0, 1, 1, 0, 0, 1, 0, …
$ rt <dbl> 0.7136441, 0.6271285, 0.6703410, 0.5738488, 0.8405913, 0.667…
$ choice <fct> right, right, left, right, right, right, right, left, left, …
$ correct <dbl> 1, 1, 0, 1, 0, 1, 0, 1, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, …
$ condition <fct> valid, valid, neutral, neutral, valid, neutral, neutral, val…Binary Choices
Pro Versuchsperson
data# A tibble: 1,440 × 9
trial ID cue direction response rt choice correct condition
<dbl> <fct> <fct> <fct> <dbl> <dbl> <fct> <dbl> <fct>
1 0 JH right right 1 0.714 right 1 valid
2 1 JH right right 1 0.627 right 1 valid
3 2 JH none right 0 0.670 left 0 neutral
4 3 JH none right 1 0.574 right 1 neutral
5 4 JH left left 1 0.841 right 0 valid
6 5 JH none right 1 0.668 right 1 neutral
7 6 JH none left 1 1.12 right 0 neutral
8 7 JH left left 0 0.640 left 1 valid
9 8 JH left right 0 1.13 left 0 invalid
10 9 JH none right 1 1.03 right 1 neutral
# … with 1,430 more rowsdata |>
group_by(ID, condition)# A tibble: 1,440 × 9
# Groups: ID, condition [27]
trial ID cue direction response rt choice correct condition
<dbl> <fct> <fct> <fct> <dbl> <dbl> <fct> <dbl> <fct>
1 0 JH right right 1 0.714 right 1 valid
2 1 JH right right 1 0.627 right 1 valid
3 2 JH none right 0 0.670 left 0 neutral
4 3 JH none right 1 0.574 right 1 neutral
5 4 JH left left 1 0.841 right 0 valid
6 5 JH none right 1 0.668 right 1 neutral
7 6 JH none left 1 1.12 right 0 neutral
8 7 JH left left 0 0.640 left 1 valid
9 8 JH left right 0 1.13 left 0 invalid
10 9 JH none right 1 1.03 right 1 neutral
# … with 1,430 more rowsaccuracy# A tibble: 27 × 5
# Groups: ID [9]
ID condition N ncorrect accuracy
<fct> <fct> <int> <dbl> <dbl>
1 JH invalid 16 13 0.812
2 JH neutral 80 66 0.825
3 JH valid 64 60 0.938
4 NS invalid 16 11 0.688
5 NS neutral 80 56 0.7
6 NS valid 64 58 0.906
7 rh invalid 16 2 0.125
8 rh neutral 80 64 0.8
9 rh valid 64 61 0.953
10 sb invalid 16 1 0.0625
# … with 17 more rowsVisualisieren
accuracy |>
ggplot(aes(x = condition, y = accuracy, fill = condition)) +
geom_col() +
geom_line(aes(group = ID), size = 2) +
geom_point(size = 8) +
scale_fill_manual(
values = c(invalid = "#9E0142",
neutral = "#C4C4B7",
valid = "#2EC762")
) +
labs(
x = "Cue",
y = "Proportion correct",
title = "Accuracy per person/condition"
) +
theme_linedraw(base_size = 28) +
facet_wrap(~ID)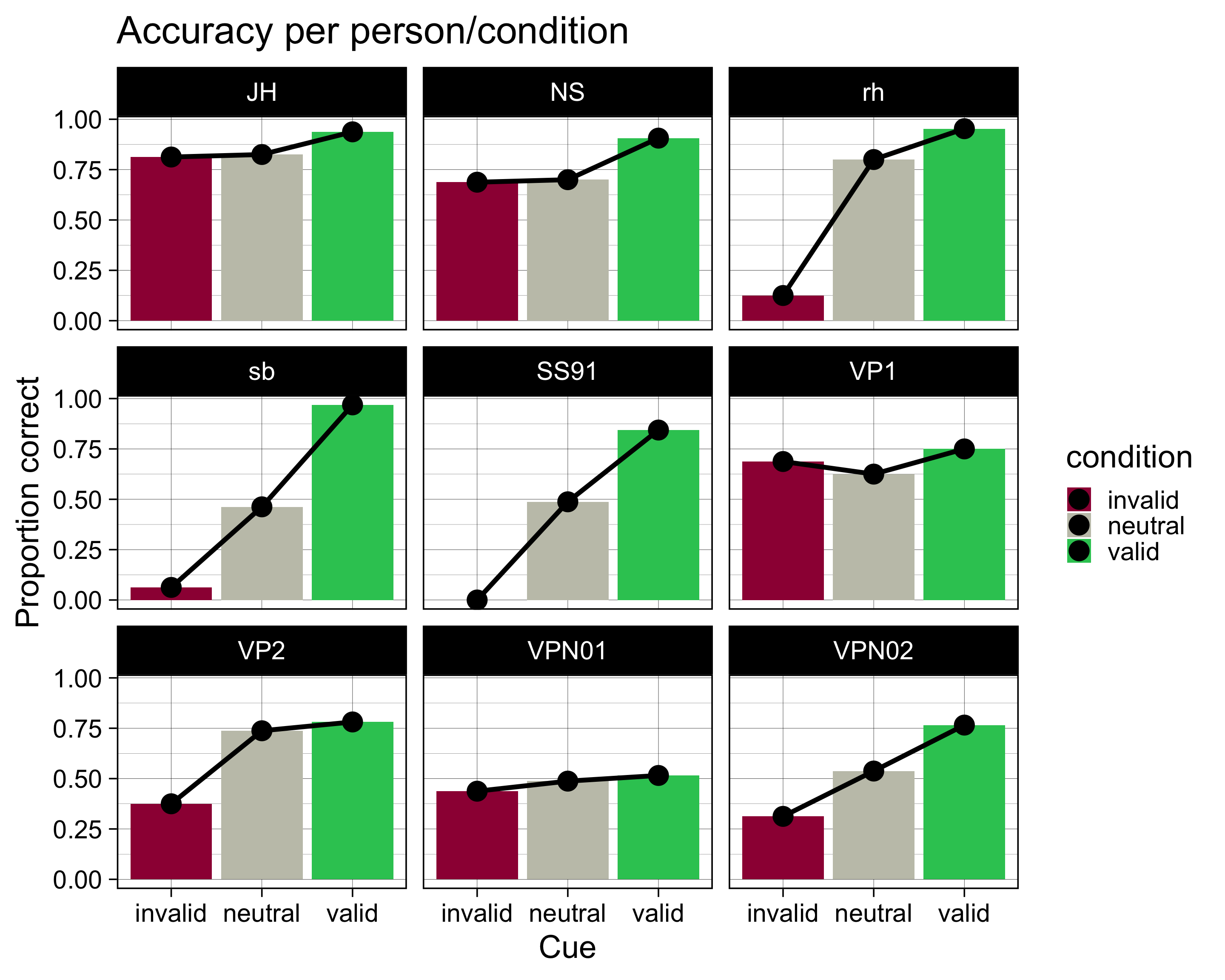
Über Versuchsperson aggregieren
Ein Exkurs über Within-person Standardfehler
dfl <- dfw |>
pivot_longer(contains("test"),
names_to = "condition",
values_to = "value") |>
mutate(condition = as_factor(condition))dflsum <- dfl |>
Rmisc::summarySEwithin(measurevar = "value",
withinvars = "condition",
idvar = "subject",
na.rm = FALSE,
conf.interval = 0.95)dflsum |>
ggplot(aes(x = condition, y = value, group = 1)) +
geom_line() +
geom_errorbar(width = 0.1, aes(ymin = value-ci, ymax = value+ci)) +
geom_point(shape = 21, size = 3, fill = "white") +
ylim(40,60)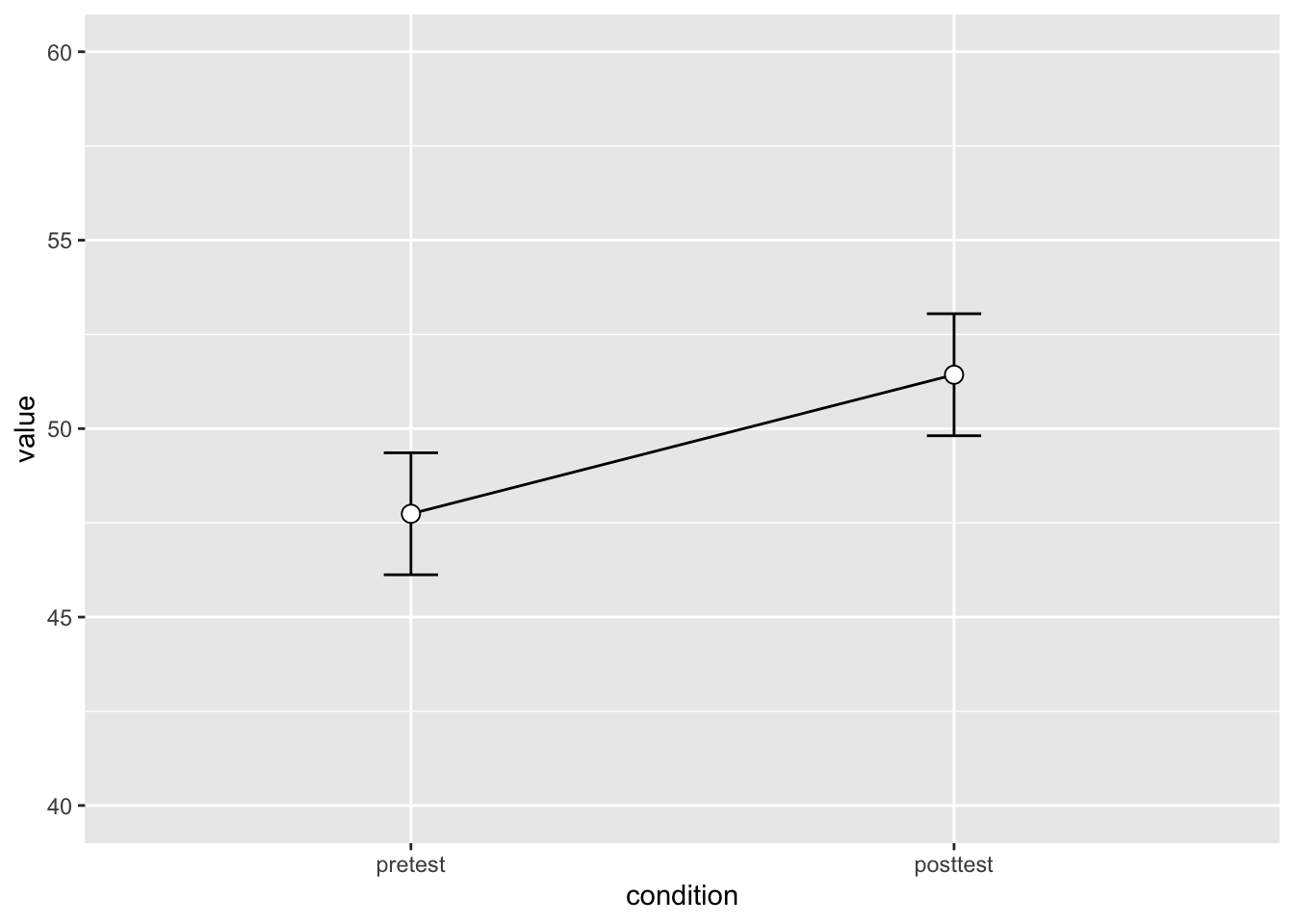
# Plot the individuals
dfl |>
ggplot(aes(x=condition, y=value, colour=subject, group=subject)) +
geom_line() + geom_point(shape=21, fill="white") +
ylim(ymin,ymax)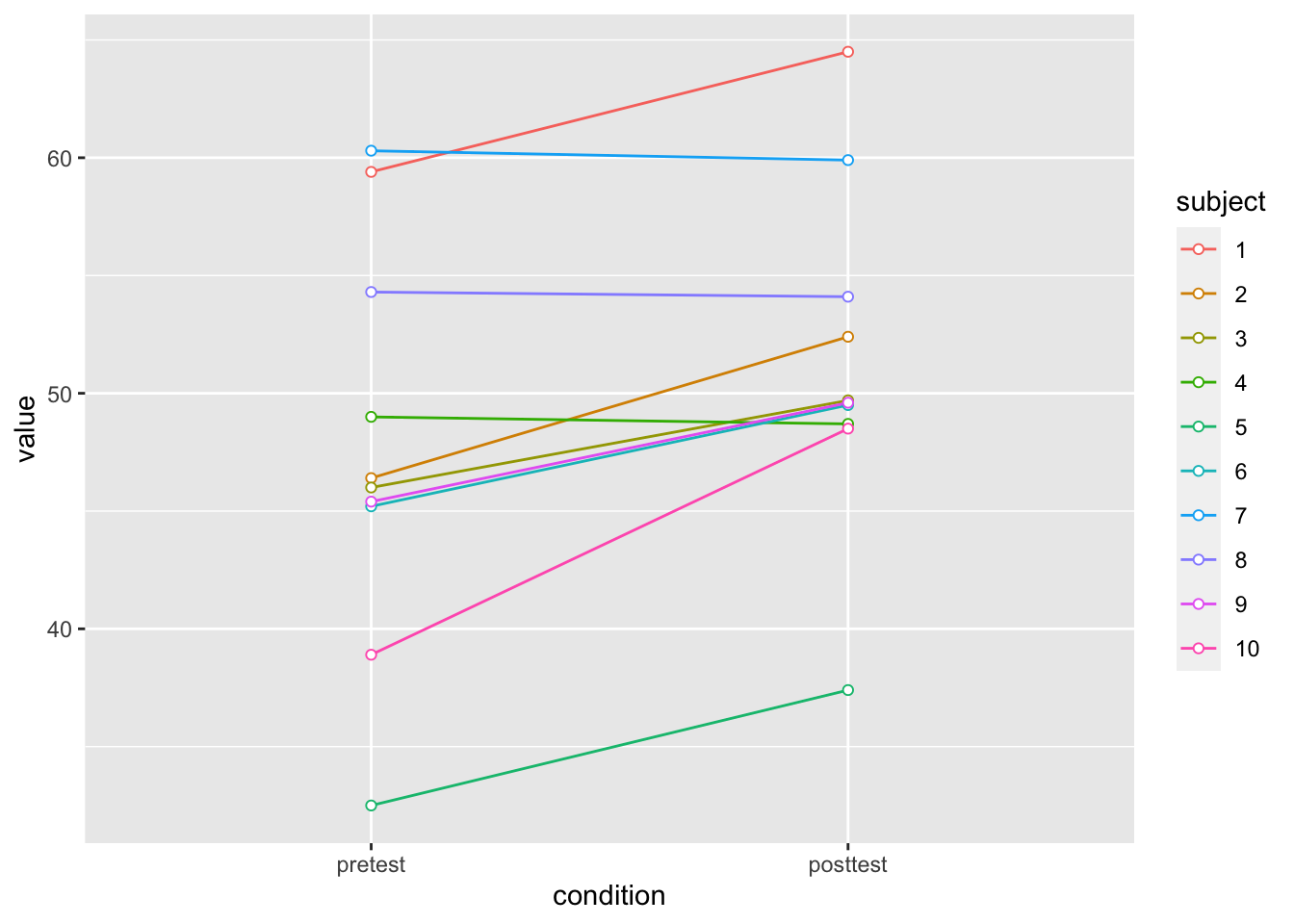
dfNorm_long <- Rmisc::normDataWithin(data=dfl, idvar="subject", measurevar="value")
?Rmisc::normDataWithin
dfNorm_long |>
ggplot(aes(x=condition, y=valueNormed, colour=subject, group=subject)) +
geom_line() + geom_point(shape=21, fill="white") +
ylim(ymin,ymax)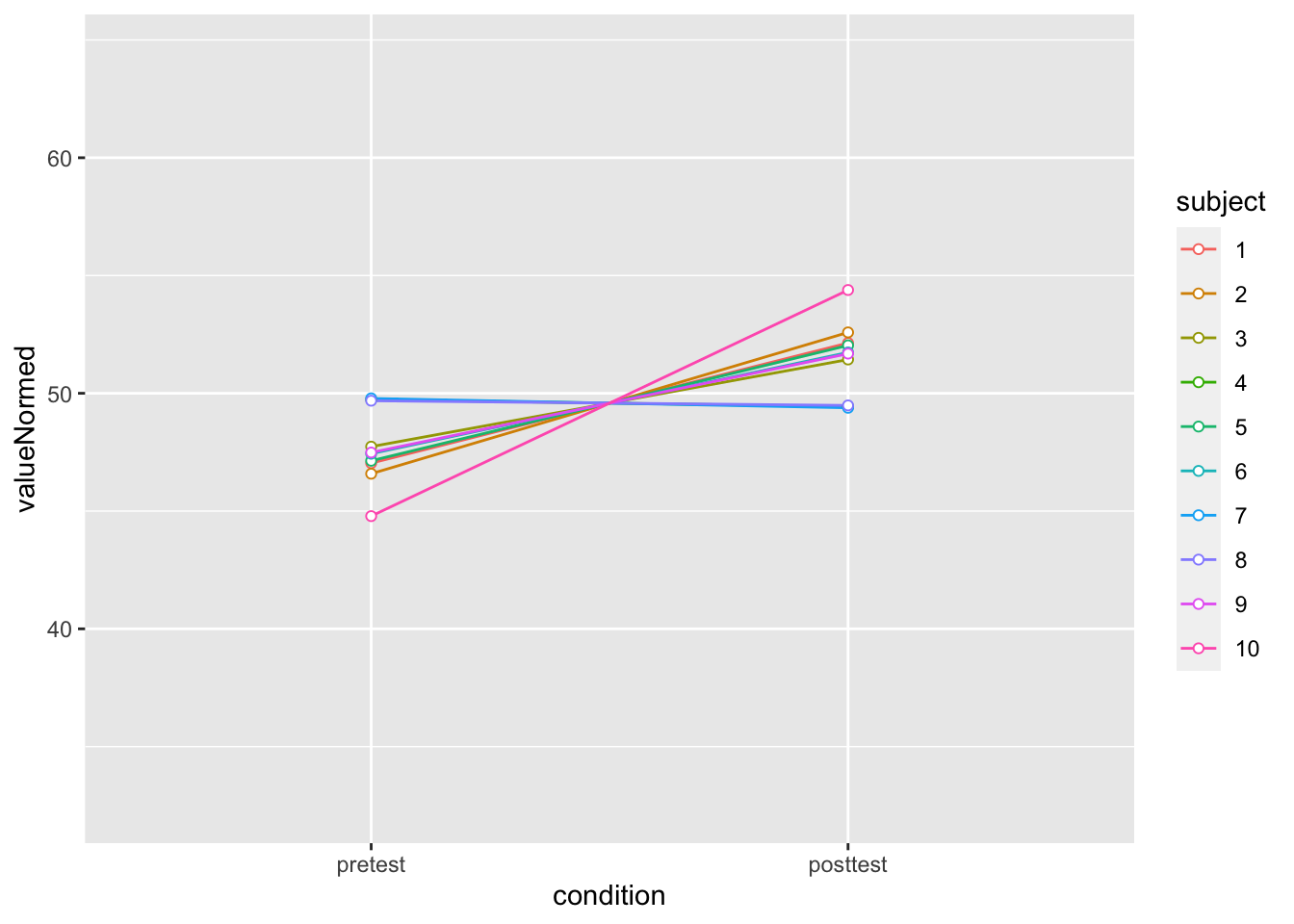
# Instead of summarySEwithin, use summarySE, which treats condition as though it were a between-subjects variable
dflsum_between <- Rmisc::summarySE(data = dfl,
measurevar = "value",
groupvars = "condition",
na.rm = FALSE,
conf.interval = .95)
dflsum_between condition N value sd se ci
1 pretest 10 47.74 8.598992 2.719240 6.151348
2 posttest 10 51.43 7.253972 2.293907 5.189179# Show the between-S CI's in red, and the within-S CI's in black
dflsum_between |>
ggplot(aes(x=condition, y=value, group=1)) +
geom_line() +
geom_errorbar(width=.1, aes(ymin=value-ci, ymax=value+ci), colour="red") +
geom_errorbar(width=.1, aes(ymin=value-ci, ymax=value+ci), data=dflsum) +
geom_point(shape=21, size=3, fill="white") +
ylim(ymin,ymax)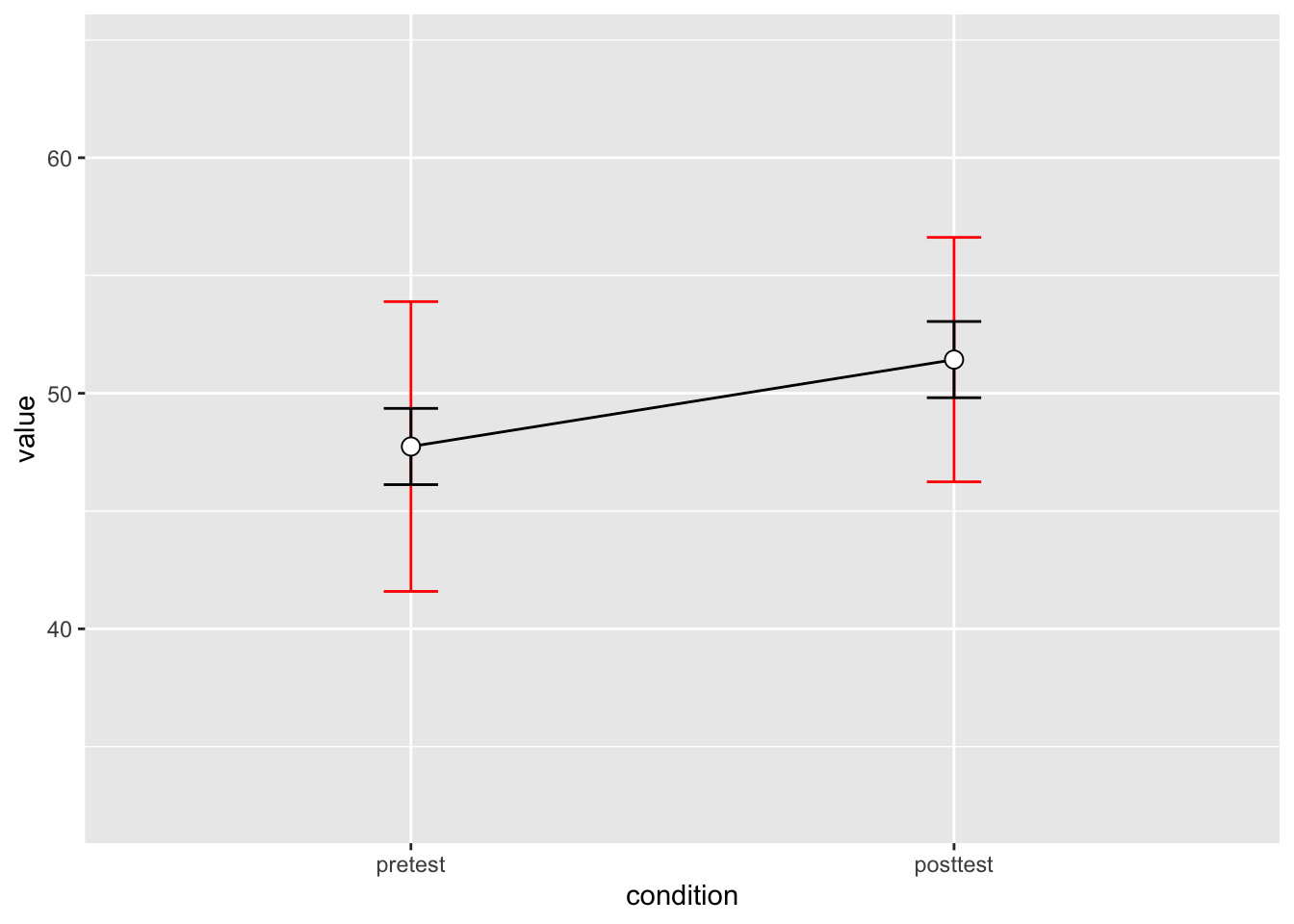
Within-person Standardfehler
accuracy |>
ggplot(aes(x = condition, y = accuracy, colour = ID, group = ID)) +
geom_line() +
geom_point(shape=21, fill="white")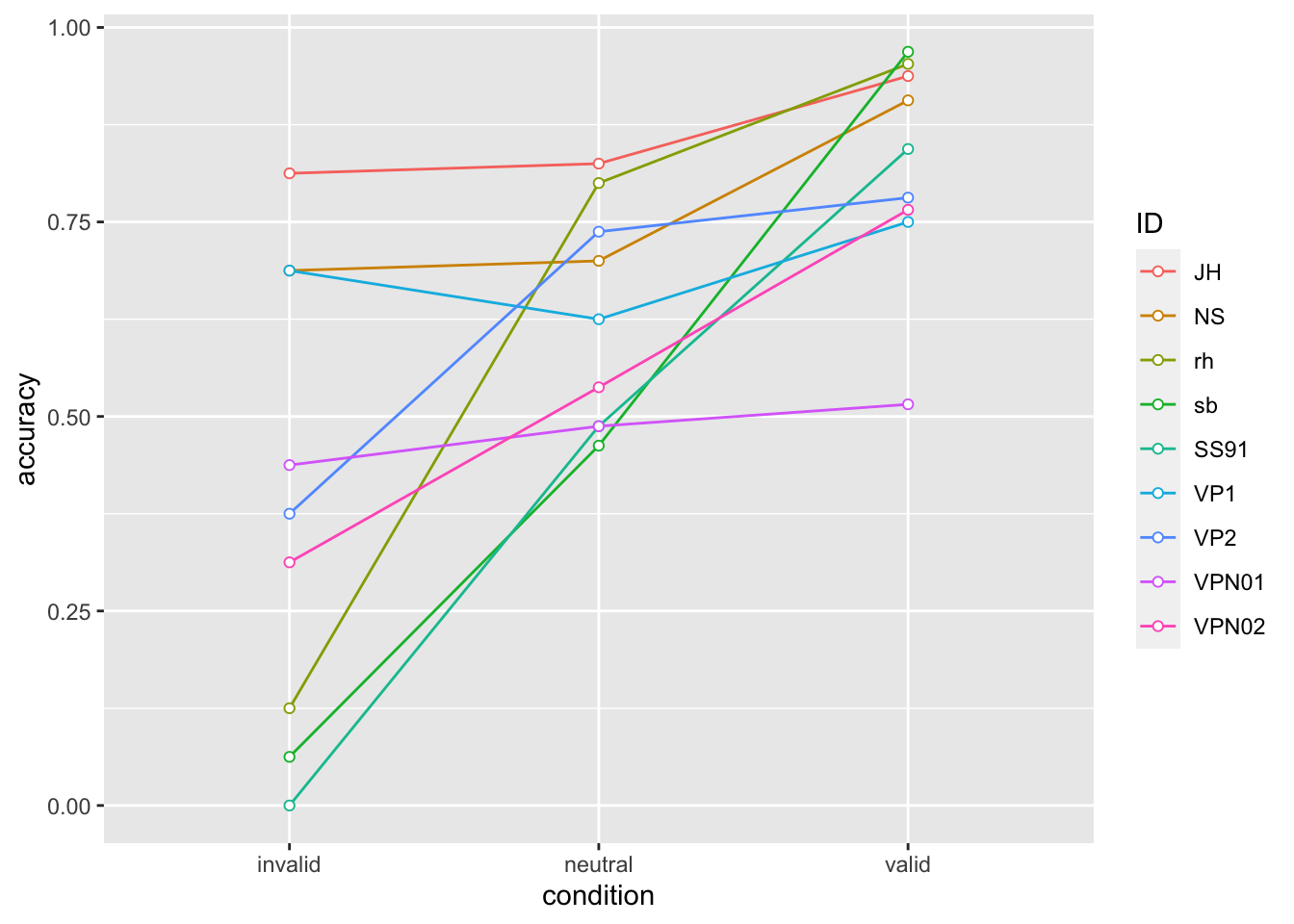
Der Standardfehler is definiert als: \[SE = sd/ \sqrt{n}\]
Leider gibt es in R keine Funktion, welche den Standardfehler berechnet (schätzt); wir können aber ganz einfach selber eine Funktion definieren.
datasum <- data |>
group_by(condition) |>
summarise(N = n(),
ccuracy = mean(correct),
sd = sd(correct),
se = se(correct))
datasum# A tibble: 3 × 5
condition N ccuracy sd se
<fct> <int> <dbl> <dbl> <dbl>
1 invalid 144 0.389 0.489 0.0408
2 neutral 720 0.629 0.483 0.0180
3 valid 576 0.825 0.381 0.0159datasum_2 <- data |>
Rmisc::summarySE(measurevar = "correct",
groupvars = "condition",
na.rm = FALSE,
conf.interval = 0.95)
datasum_2 condition N correct sd se ci
1 invalid 144 0.3888889 0.4891996 0.04076663 0.08058308
2 neutral 720 0.6291667 0.4833637 0.01801390 0.03536613
3 valid 576 0.8246528 0.3805943 0.01585810 0.03114686datasum_3 <- data |>
Rmisc::summarySEwithin(measurevar = "correct",
withinvars = "condition",
idvar = "ID",
na.rm = FALSE,
conf.interval = 0.95)
datasum_3 condition N correct sd se ci
1 invalid 144 0.3888889 0.5773528 0.04811273 0.09510406
2 neutral 720 0.6291667 0.5726512 0.02134145 0.04189901
3 valid 576 0.8246528 0.4523391 0.01884746 0.03701827p_accuracy <- datasum_3 |>
ggplot(aes(x = condition, y = correct, group = 1)) +
geom_line() +
geom_errorbar(width = .1, aes(ymin = correct-se, ymax = correct+se), colour="red") +
geom_point(shape=21, size=3, fill="white")
p_accuracy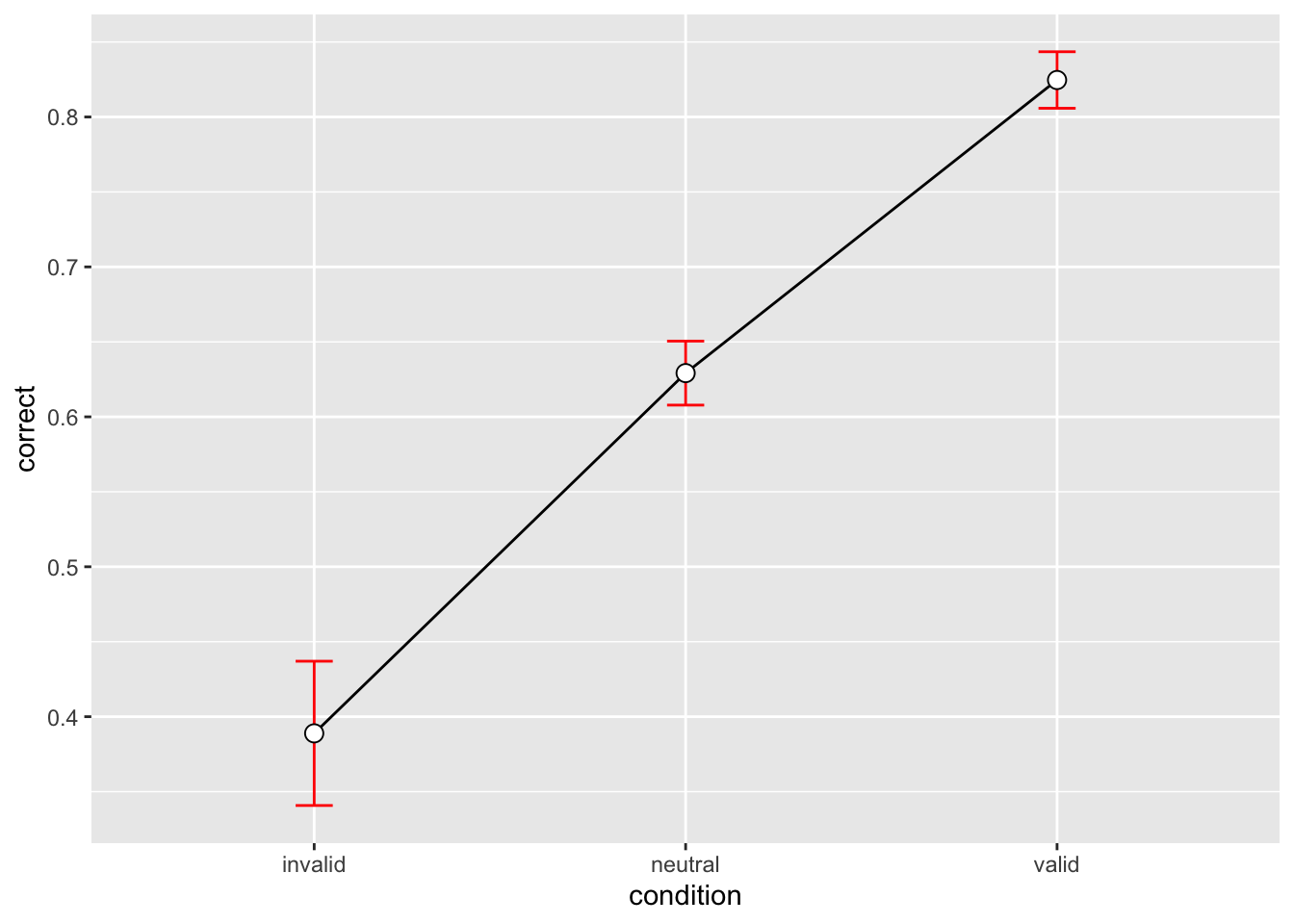
Reaktionszeiten
Pro Versuchsperson
Wir fassen die Daten pro Person pro Block mit Mittelwert, Median und Standarabweichung zusammen.
by_subj # A tibble: 27 × 5
# Groups: ID [9]
ID condition mean median sd
<fct> <fct> <dbl> <dbl> <dbl>
1 JH invalid 0.775 0.739 0.163
2 JH neutral 0.799 0.733 0.202
3 JH valid 0.696 0.658 0.190
4 NS invalid 0.894 0.913 0.207
5 NS neutral 0.885 0.844 0.201
6 NS valid 0.738 0.715 0.191
7 rh invalid 0.423 0.389 0.151
8 rh neutral 0.525 0.503 0.0841
9 rh valid 0.443 0.390 0.185
10 sb invalid 0.376 0.341 0.0924
# … with 17 more rowsEinfachere Version:
by_subj |>
ggplot(aes(x = condition, y = mean, fill = condition)) +
geom_col() +
geom_line(aes(group = ID), size = 2) +
geom_point(size = 8) +
scale_fill_manual(
values = c(invalid = "#9E0142",
neutral = "#C4C4B7",
valid = "#2EC762")
) +
labs(
x = "Cue",
y = "Response time") +
theme_linedraw(base_size = 28) +
facet_wrap(~ID)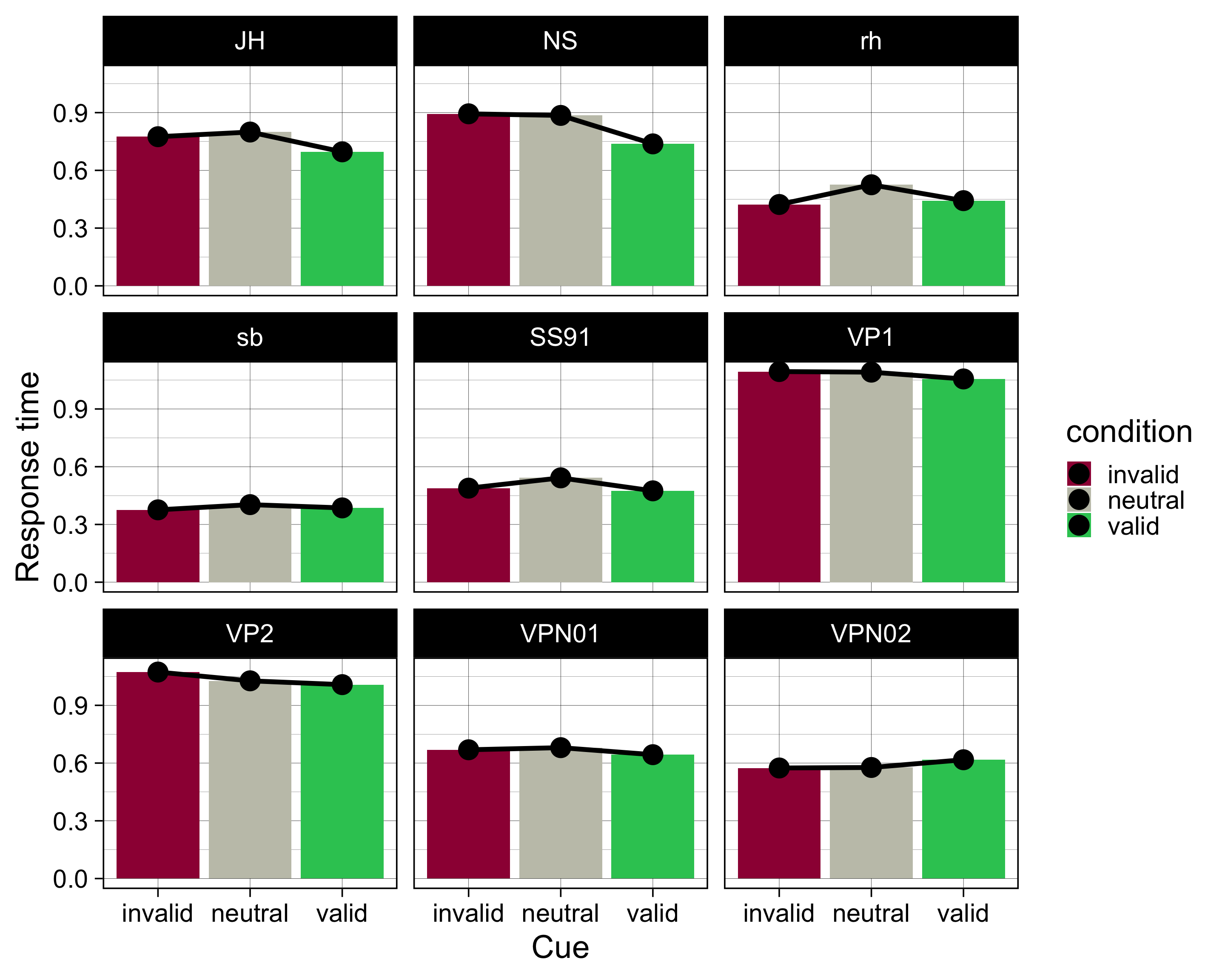
by_subj |>
ggplot(aes(condition, mean)) +
geom_line(aes(group = 1), linetype = 3) +
geom_errorbar(aes(ymin = mean-se, ymax = mean+se),
width = 0.2, size=1, color="blue") +
geom_point(size = 2) +
facet_wrap(~ID, scales = "free_y")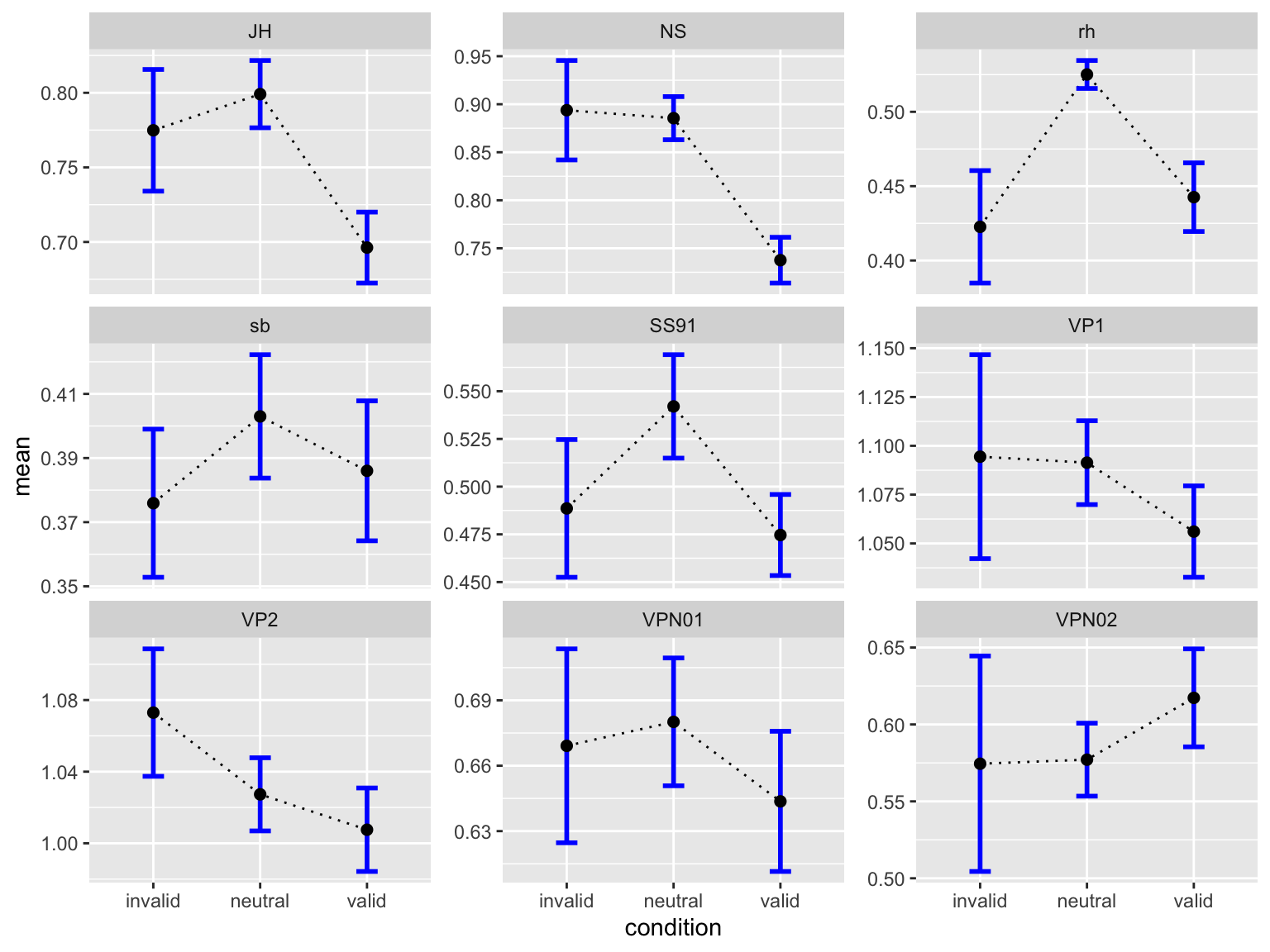
Über Versuchsperson aggregieren
rtsum <- data |>
drop_na(rt) |>
Rmisc::summarySEwithin(measurevar = "rt",
withinvars = "condition",
idvar = "ID",
na.rm = FALSE,
conf.interval = 0.95)
rtsum condition N rt sd se ci
1 invalid 141 0.7055247 0.2204498 0.01856522 0.03670444
2 neutral 710 0.7238269 0.2449543 0.00919297 0.01804870
3 valid 568 0.6716487 0.2482698 0.01041717 0.02046095p_rt <- rtsum |>
ggplot(aes(x = condition, y = rt, group = 1)) +
geom_line() +
geom_errorbar(width = .1, aes(ymin = rt-se, ymax = rt+se), colour="red") +
geom_point(shape=21, size=3, fill="white")p_rt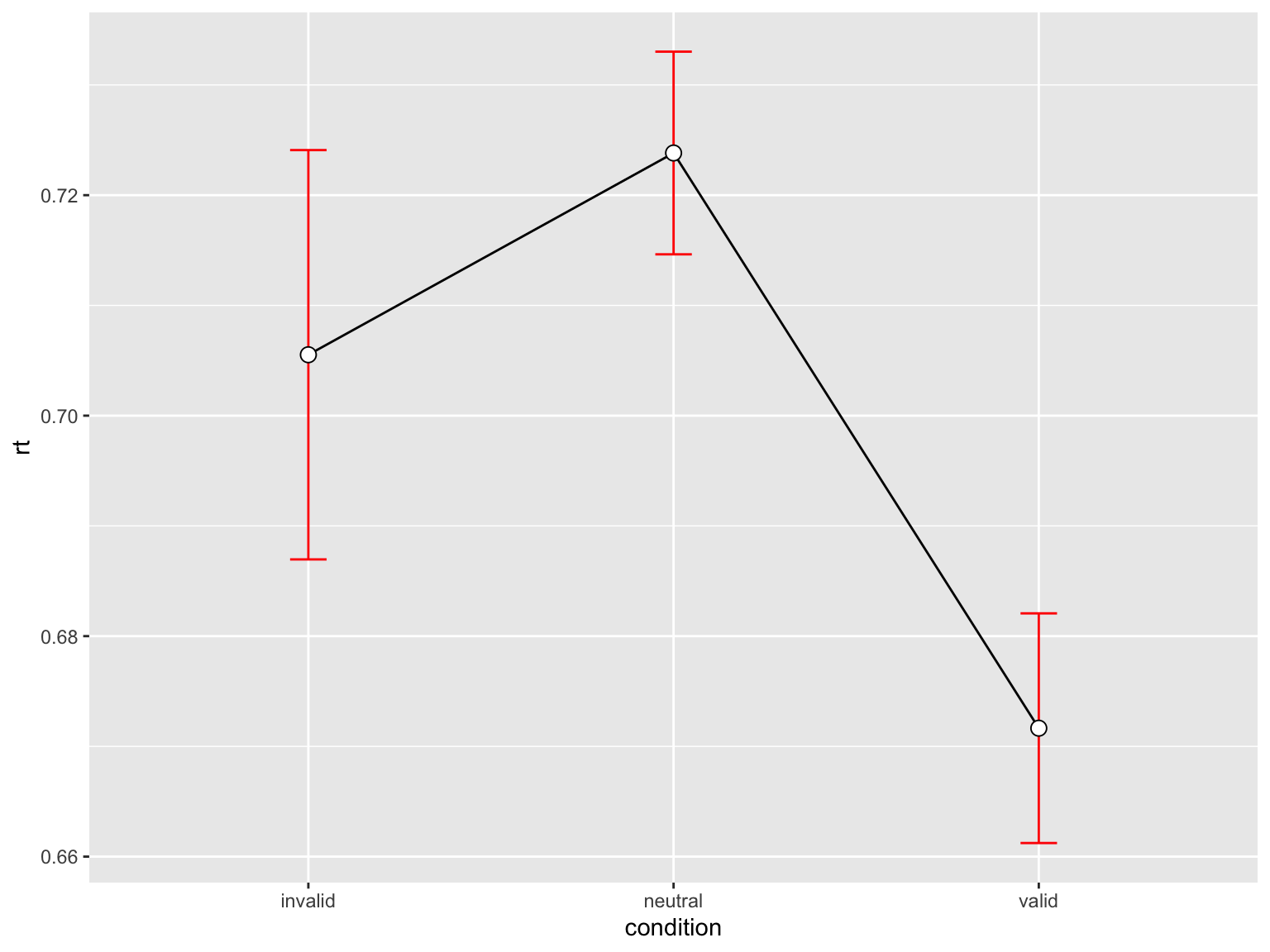
p_accuracy / p_rt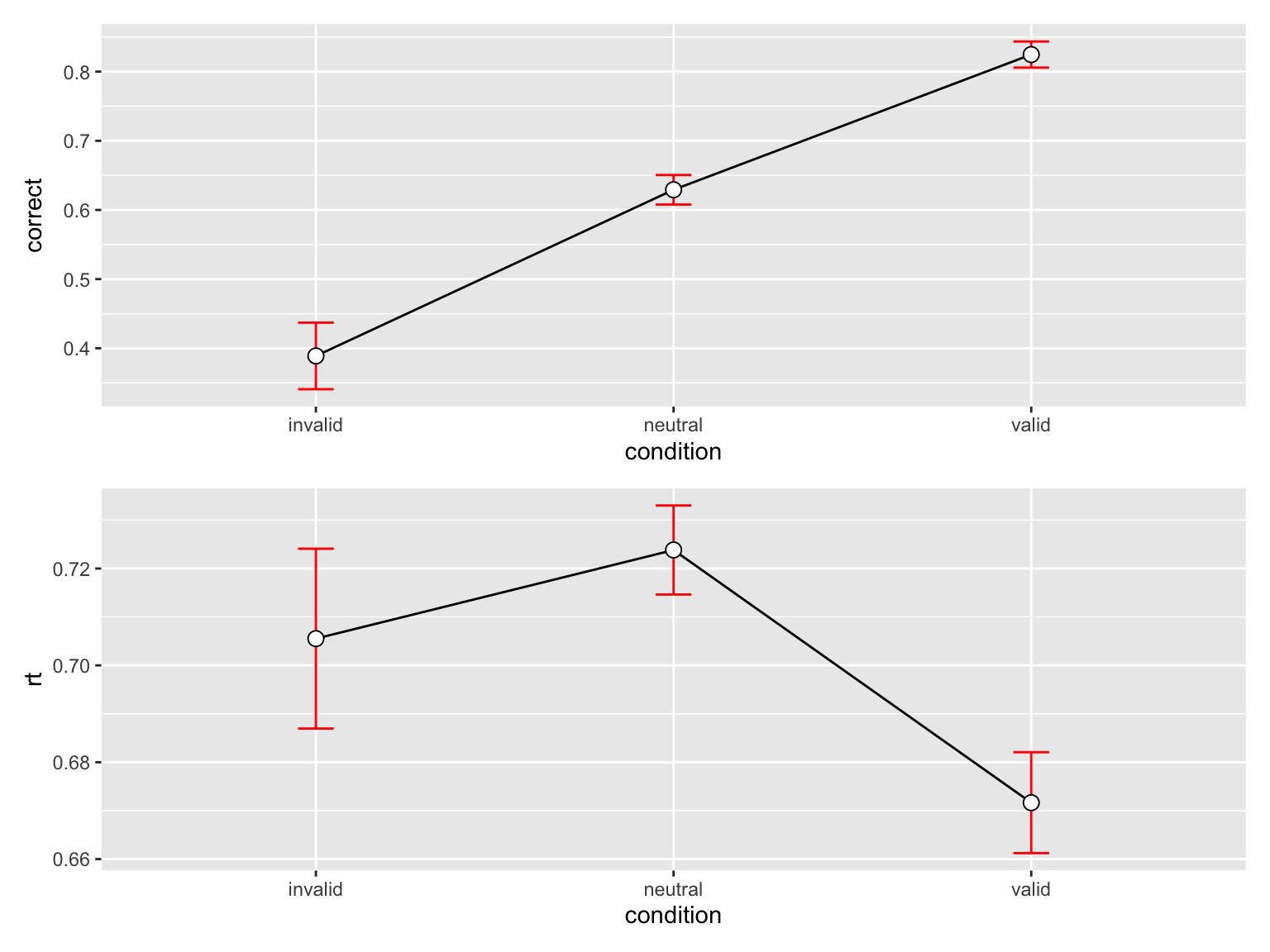
Reuse
Citation
BibTeX citation:
@online{ellis2022,
author = {Andrew Ellis},
title = {Daten Bearbeiten Und Zusammenfassen},
date = {2022-03-15},
url = {https://kogpsy.github.io/neuroscicomplabFS22//pages/chapters/04_summarizing_data.html},
langid = {en}
}
For attribution, please cite this work as:
Andrew Ellis. 2022. “Daten Bearbeiten Und Zusammenfassen.”
March 15, 2022. https://kogpsy.github.io/neuroscicomplabFS22//pages/chapters/04_summarizing_data.html.